Call:
lm(formula = Hg ~ size, data = HgDat_df)
Residuals:
Min 1Q Median 3Q Max
-0.28712 -0.13507 -0.05476 0.10644 0.35471
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.781644 0.084717 9.227 1.98e-13 ***
size 0.020017 0.002067 9.682 3.16e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1874 on 65 degrees of freedom
Multiple R-squared: 0.5905, Adjusted R-squared: 0.5843
F-statistic: 93.75 on 1 and 65 DF, p-value: 3.165e-14Mixed Models: part two
Announcements
Mixed Effects Models this week (lab) –> One “lab” with me
Next week: Generalized Mixed Effects Models (No Lab)
Next week (Friday): Work on your plot
Back to our example
Back to our example
We set 4 nets
Sites were randomly chosen
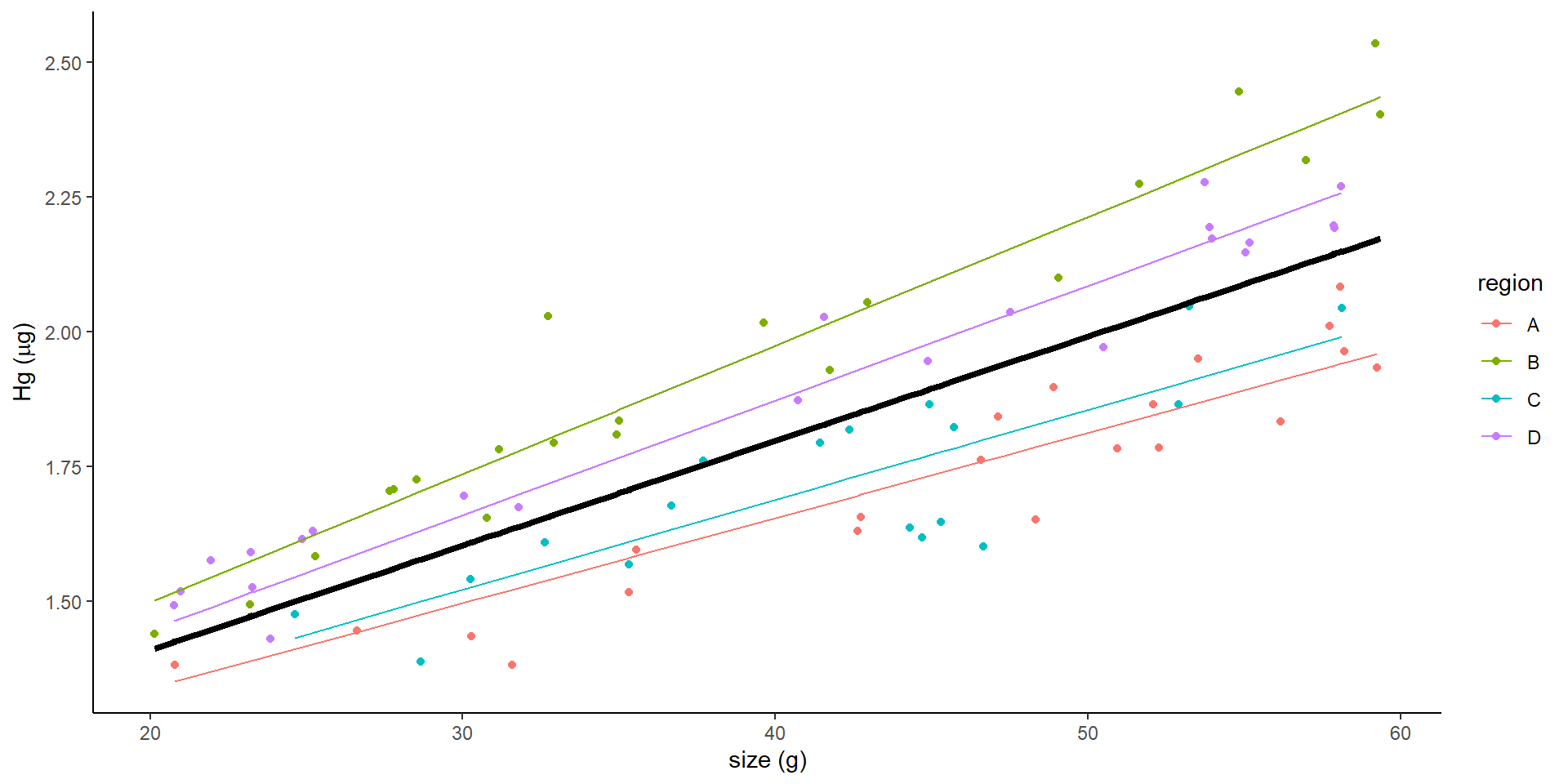
We are interested in the content of Hg in Walleye
Hg is correlated with size. The bigger the fish, the higher Hg content it has
So, normally we could do one thing:
\(E[Hg_i] = \beta_0 + \beta_1 \times size_{i}\)
\(y_i \sim N(mean = E[Hg_i], \sigma)\)
Linear model
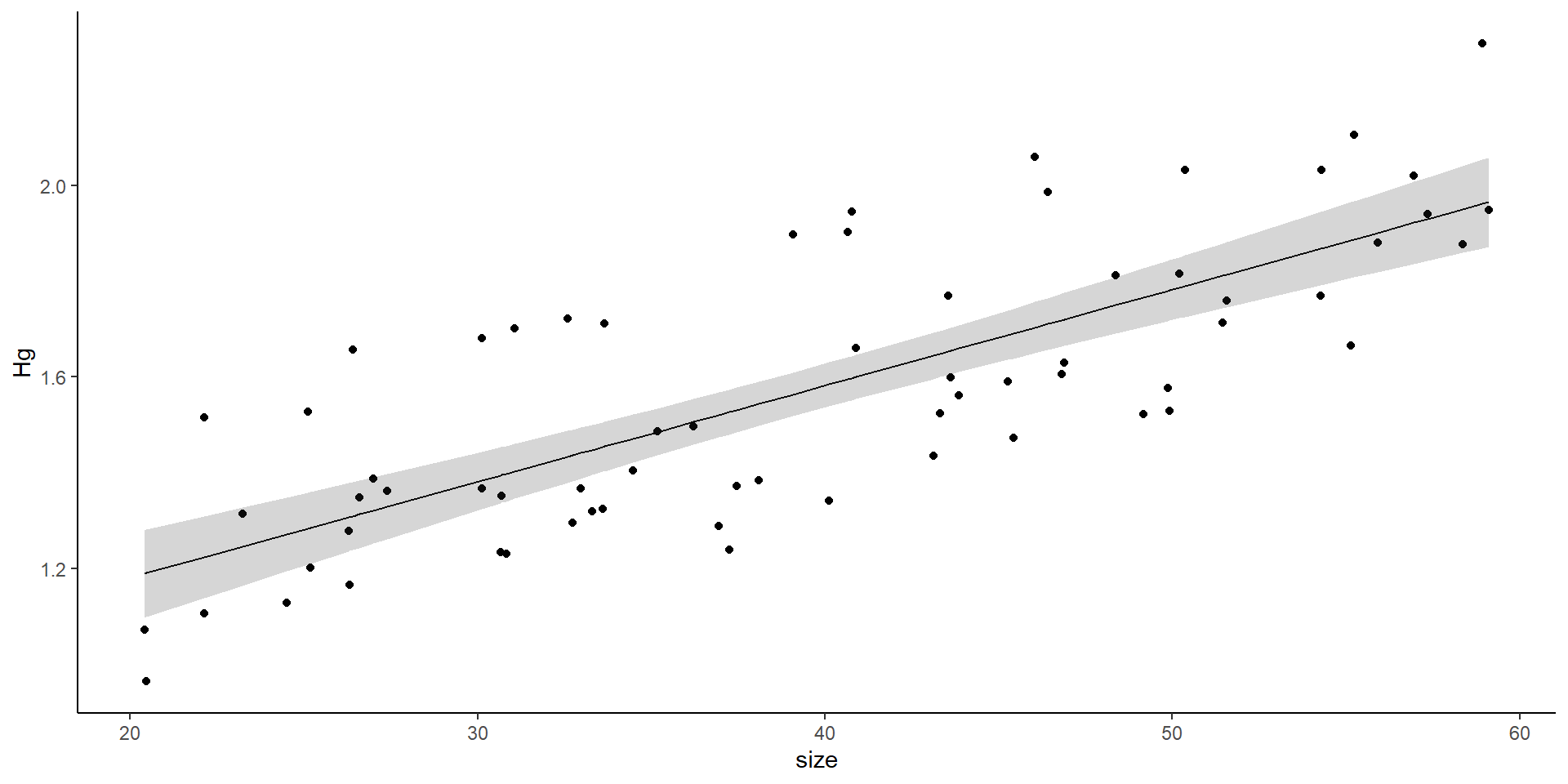
Linear model
Use coefficients to estimate size at over 1.1 \(\mu g\) .
Limit fish of that size
Assumptions of a Linear Model
Normality
Homoschedasticity
Linearity
Independence –> of data
Independence –> of error (or residuals)
Linear Model
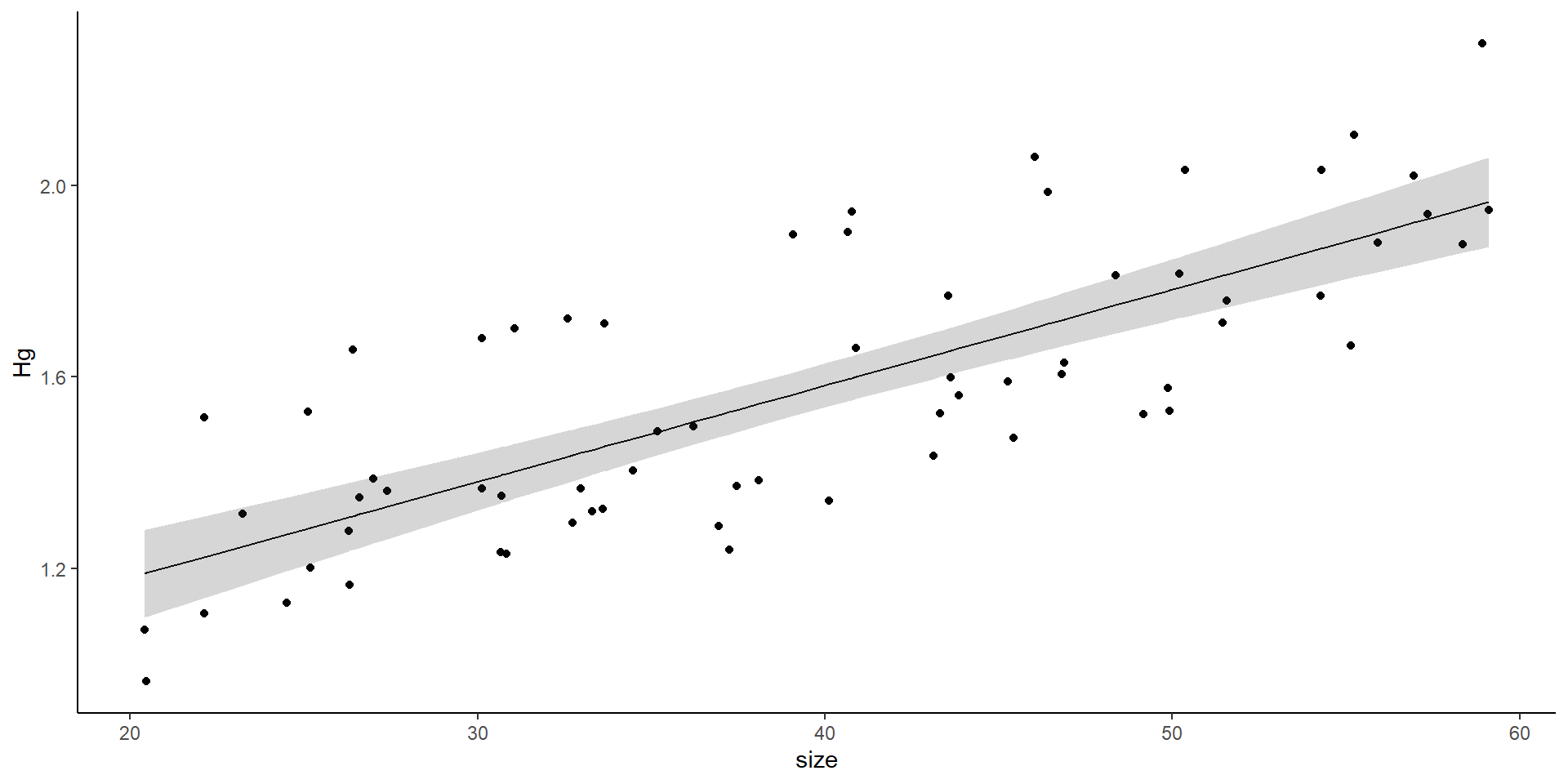
If we plot the “sites”?
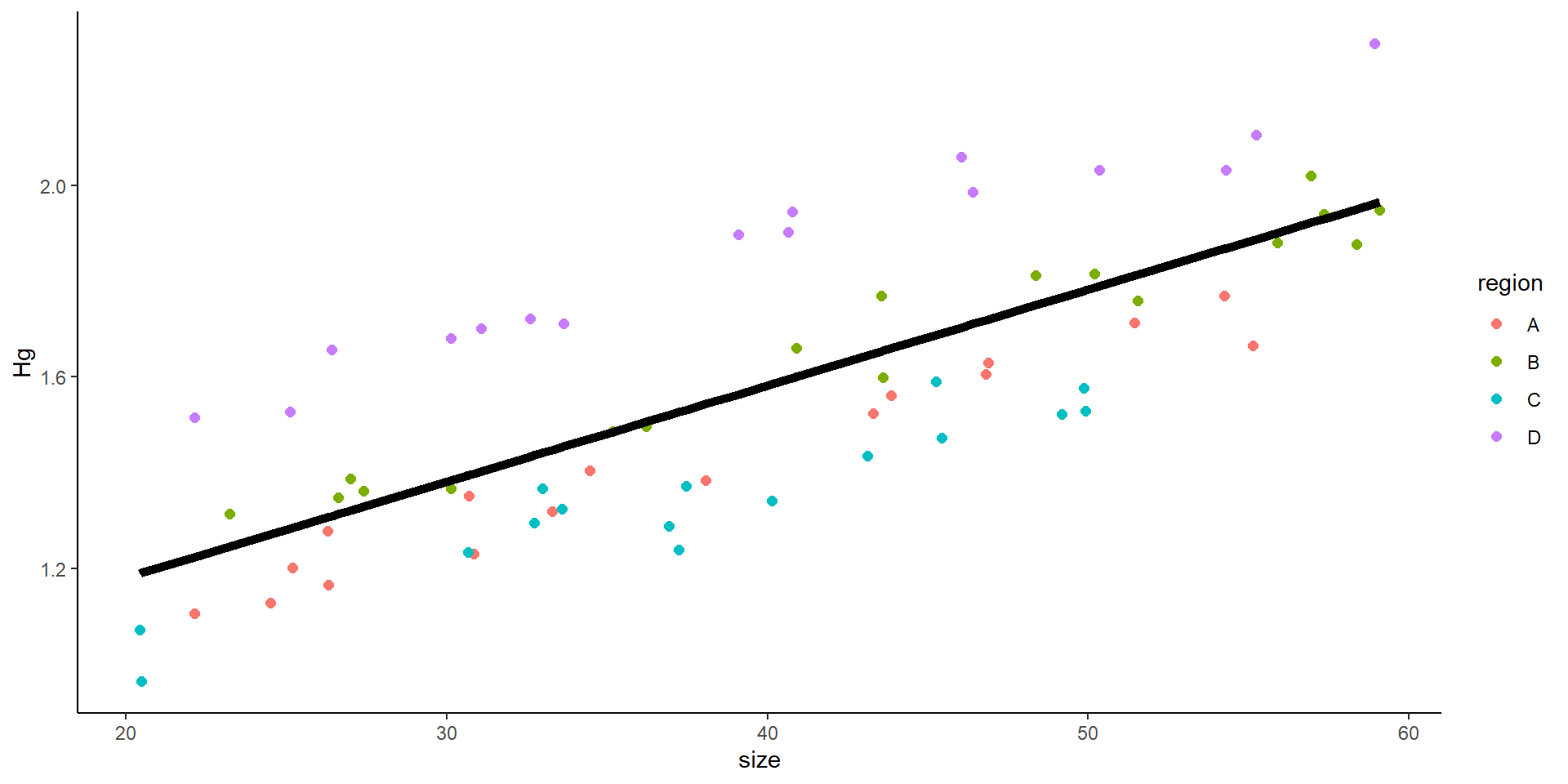
The variance can affect the slope, the intercept, or both
Random effects introduce variance.
It can introduce variance to the intercept or to the slope
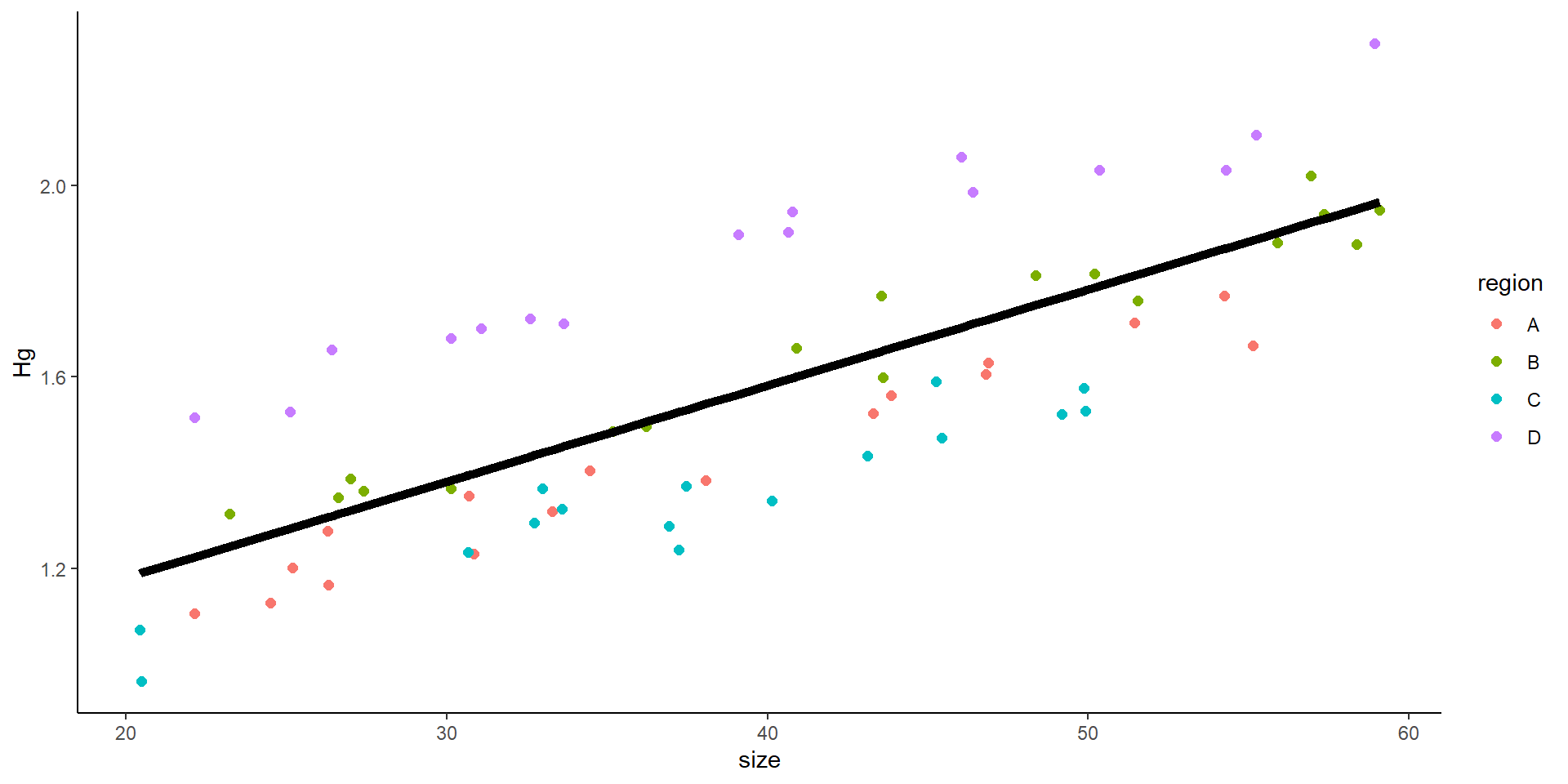
Mixed model
No mixed model:
\[ Hg_i \sim \beta_0 + \beta_1size_{i} + \epsilon_i \]
What is i?
We have two sources of variance (error): The fish (i) and the net (j)
i individuals, j sites (4)
Mixed model
No mixed model:
\[Hg_i \sim \beta_0 + \beta_1size_{i} + \epsilon_i \]
Mixed model: i individuals and j sites introduced as error.
We can do a mixed model with mixed intercept or mixed slope
mixed intercept (nets only affect the intercept) –> analogous to additive model:
\[ Hg_{ij} \sim \underbrace{(\beta_0 +\underbrace{\gamma_j}_{\text{Random intercept}})}_{intercept} + \underbrace{\beta_1size_{i}}_{slope} +\underbrace{\epsilon_i}_\text{ind var} \]
Variance comes from random “selection” of fish within a net
\(\epsilon_i \sim N(0, \sigma)\)
Variance comes from random “selection” of sites
\(\gamma_j \sim N(0,\sigma_j)\)
What does a mixed intercept mean?
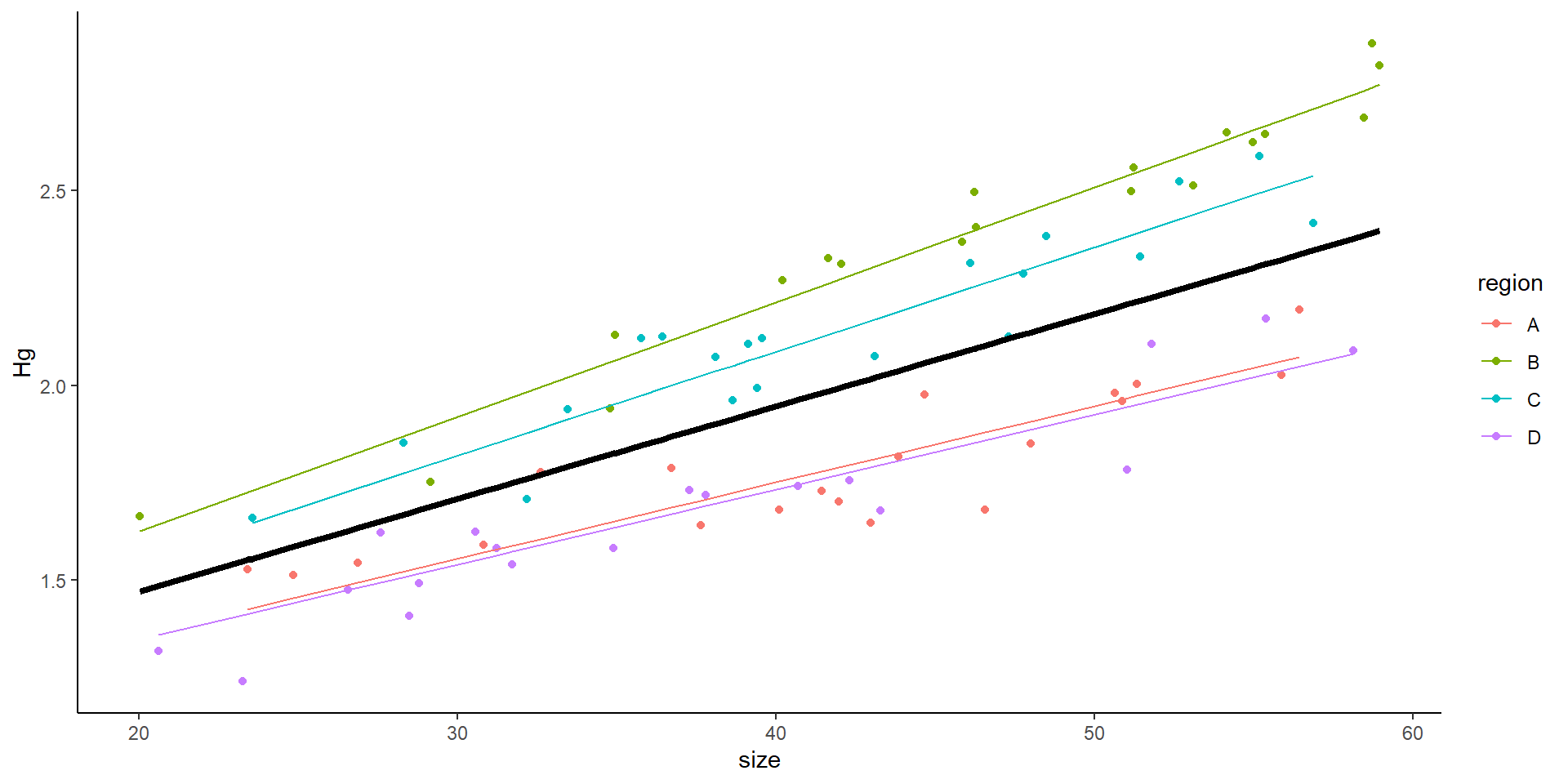
Mixed model with random intercept
This is the result of a mixed model:
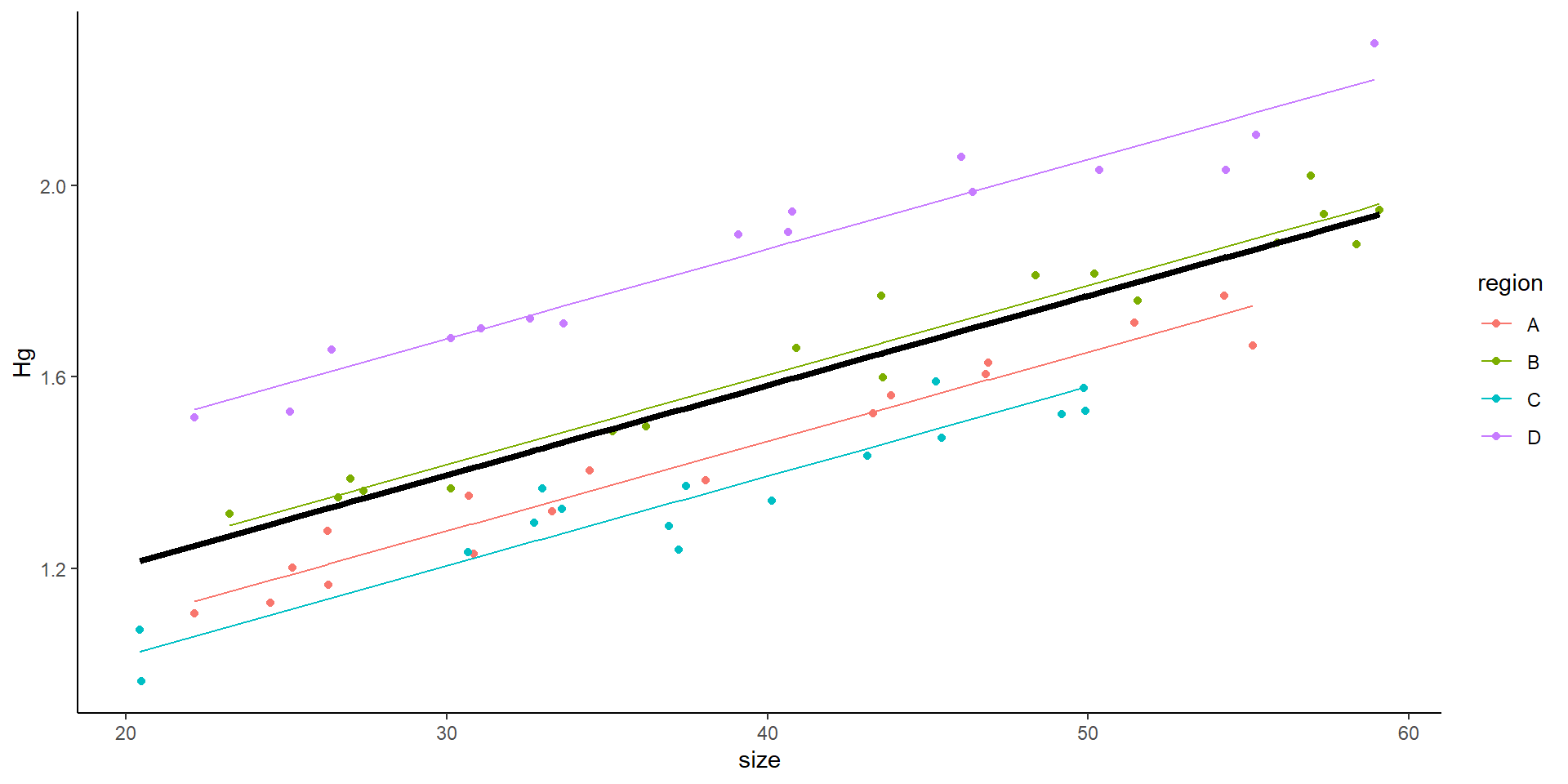
- What does the random intercept mean?
- Why not simply do:
- \(Hg_i \sim \beta_0 + \beta_1size_{i} + \beta_2rB_{i} + \beta_3rC_{i} + \beta_4rD_{i}+ \epsilon_i\)
- Model with effect of site
Mixed model with random intercept
Family: gaussian ( identity )
Formula: Hg ~ size + (1 | region)
Data: HgDat_df
AIC BIC logLik deviance df.resid
-177.1 -168.3 92.6 -185.1 63
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
region (Intercept) 0.032847 0.18124
Residual 0.002688 0.05184
Number of obs: 67, groups: region, 4
Dispersion estimate for gaussian family (sigma^2): 0.00269
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.8357038 0.0937058 8.92 <2e-16 ***
size 0.0186742 0.0005839 31.98 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1\[ Hg_{ij} = \underbrace{(\beta_0 +\underbrace{\gamma_j}_{\text{Random intercept}})}_{intercept} + \underbrace{\beta_1size_{i}}_{slope} +\underbrace{\epsilon_i}_\text{ind var} \]
Where:
\[ \epsilon_i \sim N(0,\sigma^2) \]
\[ \gamma_j \sim N(0,\sigma^2_\gamma) \]
Mixed effects models: how to run them?
Random effects are specified as \(x|g\)
x is an effect
g is grouping factor (categorical)
\(1|region\)
Effect -> 1 (intercept)
Grouping factor -> region
Mixed effects model output
Family: gaussian ( identity )
Formula: Hg ~ size + (1 | region)
Data: HgDat_df
AIC BIC logLik deviance df.resid
-177.1 -168.3 92.6 -185.1 63
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
region (Intercept) 0.032847 0.18124
Residual 0.002688 0.05184
Number of obs: 67, groups: region, 4
Dispersion estimate for gaussian family (sigma^2): 0.00269
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.8357038 0.0937058 8.92 <2e-16 ***
size 0.0186742 0.0005839 31.98 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1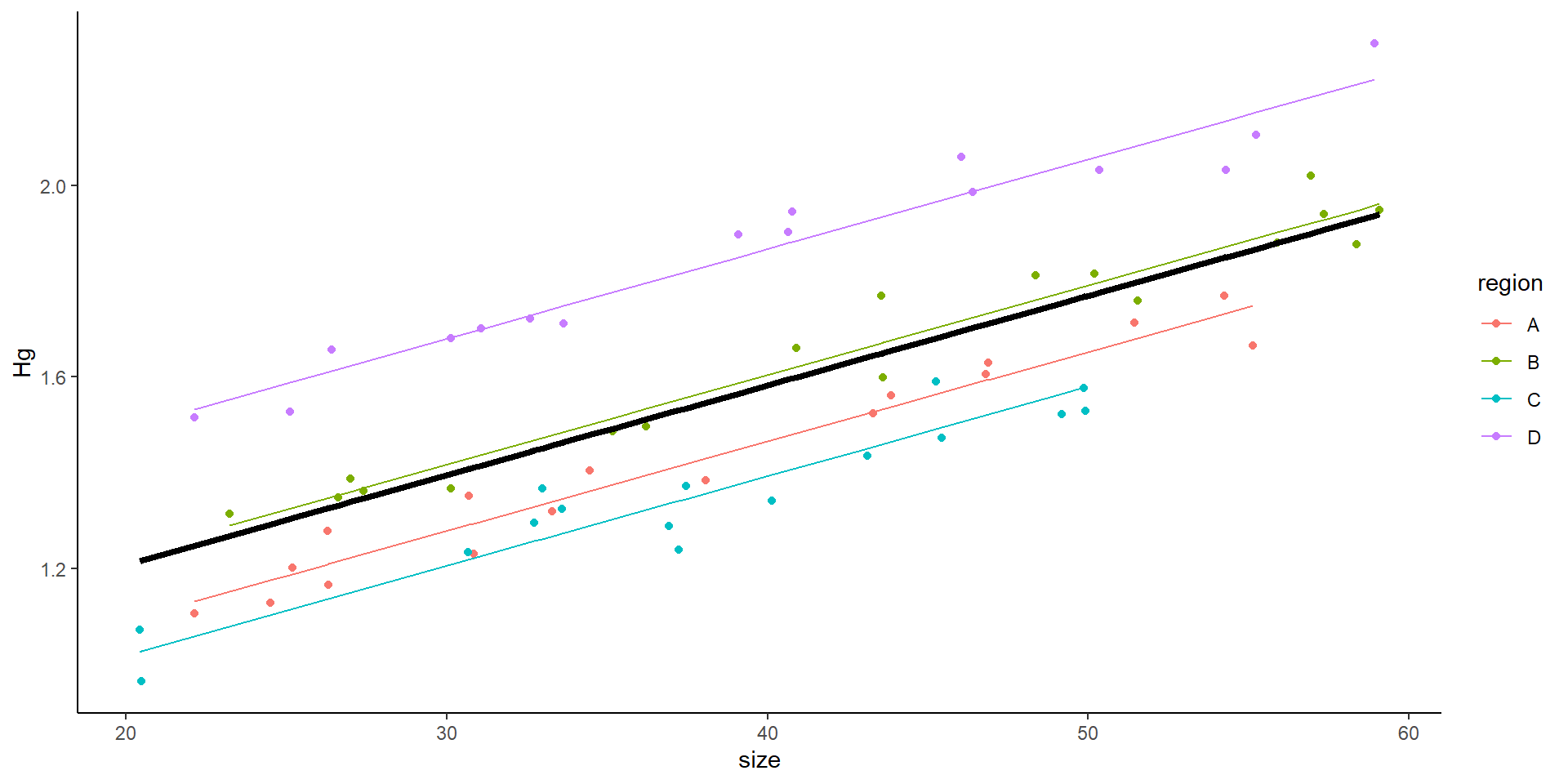
Mixed model with random intercept and random slope
\[ Hg_i \sim \beta_0 + \beta_1size_{i} + \epsilon_i \]
i individuals, j sites (4)
\[ Hg_{ij} \sim \underbrace{(\beta_0 +\underbrace{\gamma_j}_{\text{Random intercept}})}_{intercept} + \underbrace{(\beta_1+\underbrace{\psi_j}_{\text{Random slope}})size_{i}}_{slope} +\underbrace{\epsilon_i}_\text{ind var} \]
Variance comes from random “selection” of fish within a net
Variance comes from random “selection” of sites
Where,
\[ \epsilon_i \sim N(0,\sigma^2) \]
\[ \gamma_j \sim N(0,\sigma^2_\gamma) \]
\[ \psi_j \sim N(0,\sigma^2_\psi) \]
Random slope and intercept
Family: gaussian ( identity )
Formula: Hg ~ size + (1 + size | region)
Data: HgDat2_df
AIC BIC logLik deviance df.resid
-153.4 -138.4 82.7 -165.4 83
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
region (Intercept) 2.765e-02 0.166290
size 8.865e-05 0.009415 0.30
Residual 6.287e-03 0.079291
Number of obs: 89, groups: region, 4
Dispersion estimate for gaussian family (sigma^2): 0.00629
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.157873 0.089296 12.967 < 2e-16 ***
size 0.034451 0.004772 7.219 5.23e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Random effects are specified as \(x|g\)
x is an effect
g is grouping factor (categorical)
\(1 + size|region\)
Effect -> 1 (intercept) + size
Grouping factor -> region
Random slope and intercept
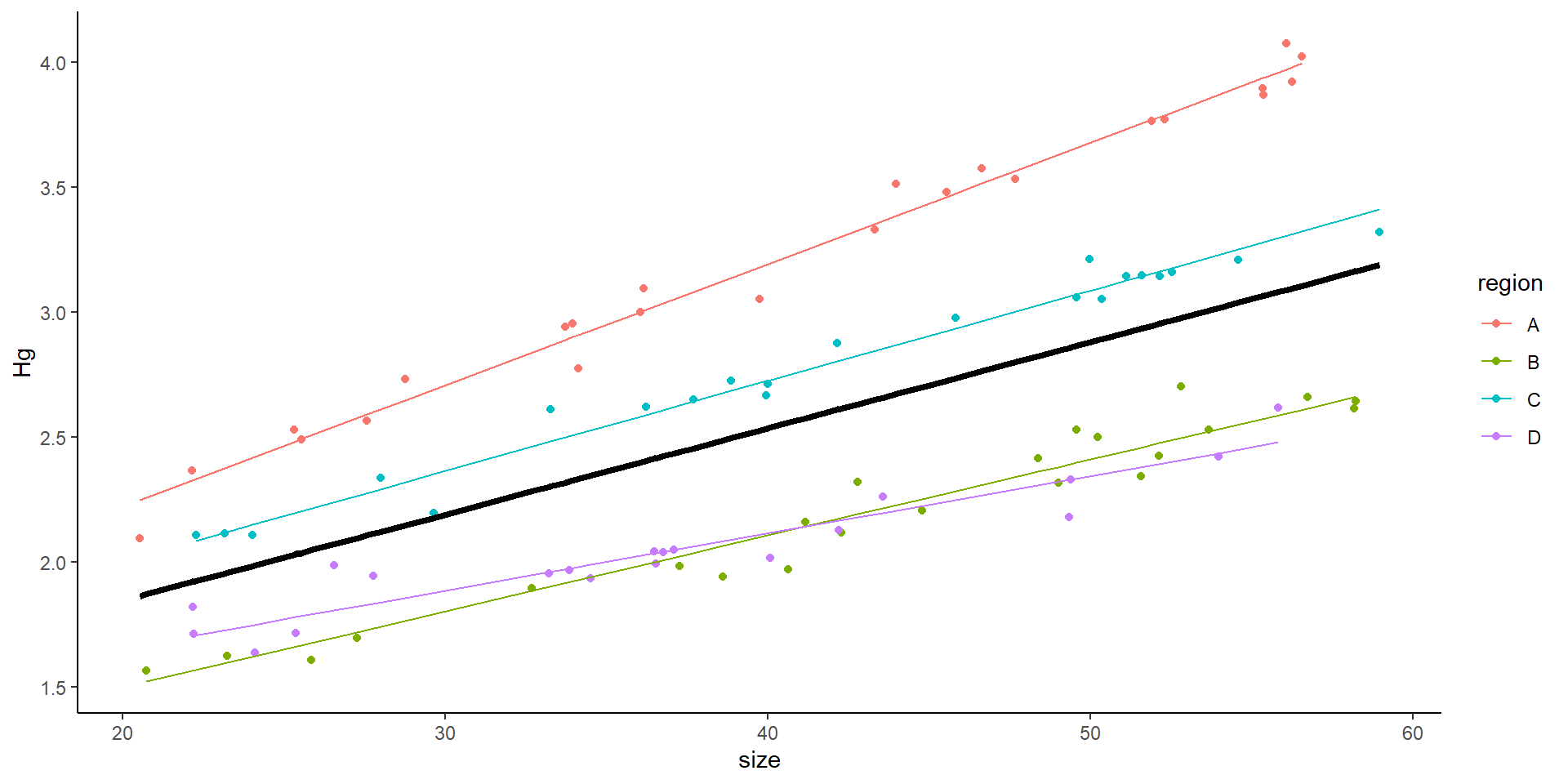
Only random slope
Family: gaussian ( identity )
Formula: Hg ~ size + (0 + size | region)
Data: HgDat2_df
AIC BIC logLik deviance df.resid
-158.4 -148.8 83.2 -166.4 79
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
region size 1.103e-05 0.003321
Residual 6.441e-03 0.080256
Number of obs: 83, groups: region, 4
Dispersion estimate for gaussian family (sigma^2): 0.00644
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.021430 0.031578 32.35 <2e-16 ***
size 0.019400 0.001817 10.68 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1We don’t do this
\(E(Hg_i) = \beta_0 + \beta_1*size_{i}\)
Pseudoreplication
\(E(Hg_i) = \beta_0 + \beta_1size_{i} + \beta_2 site2_i + \beta_3 site3_i + \beta_4 site4_i\)
We don’t care about the specific sites… we want whole population-wide!
What do we want to know?
Mercury concentration population-wide
Variance introduces by placement of the nets
\[ Hg_{ij} \sim \underbrace{(\beta_0 +\underbrace{\gamma_j}_{\text{Random intercept}})}_{intercept} + \underbrace{(\beta_1+\underbrace{\psi_j}_{\text{Random slope}})size_{i}}_{slope} +\underbrace{\epsilon}_\text{ind var} \]
\(\gamma_j \sim Normal(0,\sigma_\gamma)\)
\(\psi_j \sim Normal(0,\sigma_\psi)\)
\(\epsilon \sim Normal(0,\sigma)\)
GLM’s
- How many assumptions does it break?
Glm’s
What do Glm’s do?
- Transform the response to linear
- Have a different distribution of the residuals
If normal:
\[ \underbrace{E[y_i]}_{\text{expected value}} = \underbrace{\beta_0 + \beta_1x_{1,i} + ... \beta_mx_{m,i}}_{deterministic} \]
\[ y_i \sim \underbrace{N(mean=E[y_i], var=\sigma^2)}_{stochastic} \]
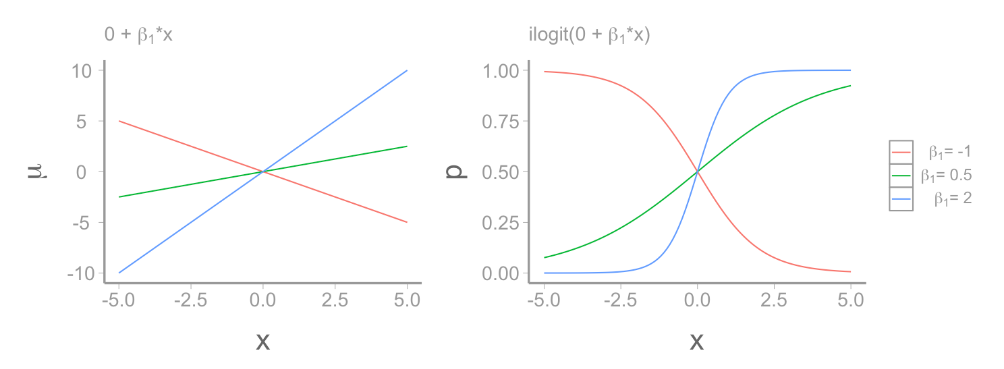
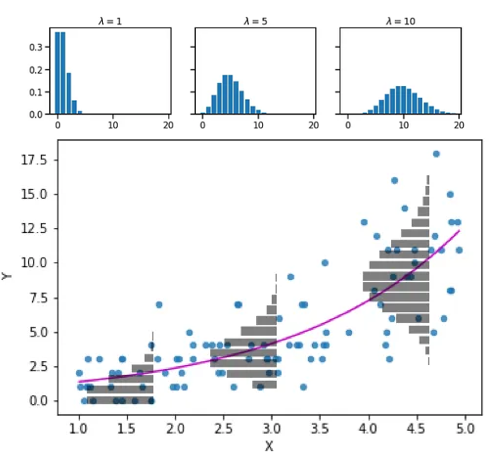
Poisson glm:
\[ \underbrace{log(\lambda)}_{\text{link function}} = \underbrace{\beta_0 + \beta_1x_{1,i} + ... \beta_mx_{m,i}}_{deterministic} \]
\[ y_i \sim \underbrace{Poisson(\lambda)}_{stochastic} \]
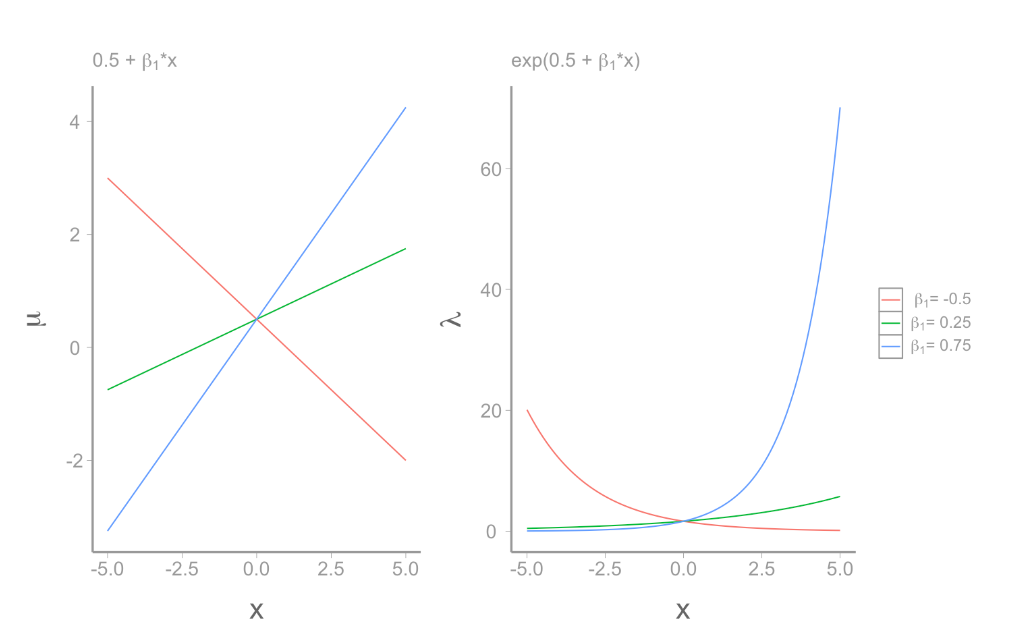
Negative Binomial glm:
\[ \underbrace{log(\lambda)}_{\text{link function}} = \underbrace{\beta_0 + \beta_1x_{1,i} + ... \beta_mx_{m,i}}_{deterministic} \]
\[ y_i \sim \underbrace{NB(\mu,\theta)}_{stochastic} \]
\(variance = \frac{\theta}{\mu+\theta}\)
GLM’s
GLM’s
GLM’s
GLM’s
Mixed models
If we set all random effects to zero, we get population mean, and predicted value for a random individual
Mixed effects

Set nets on each of those sites
- Measure 50, 43, 67, and 90 fish. For number of parasites OR presence of parasites?
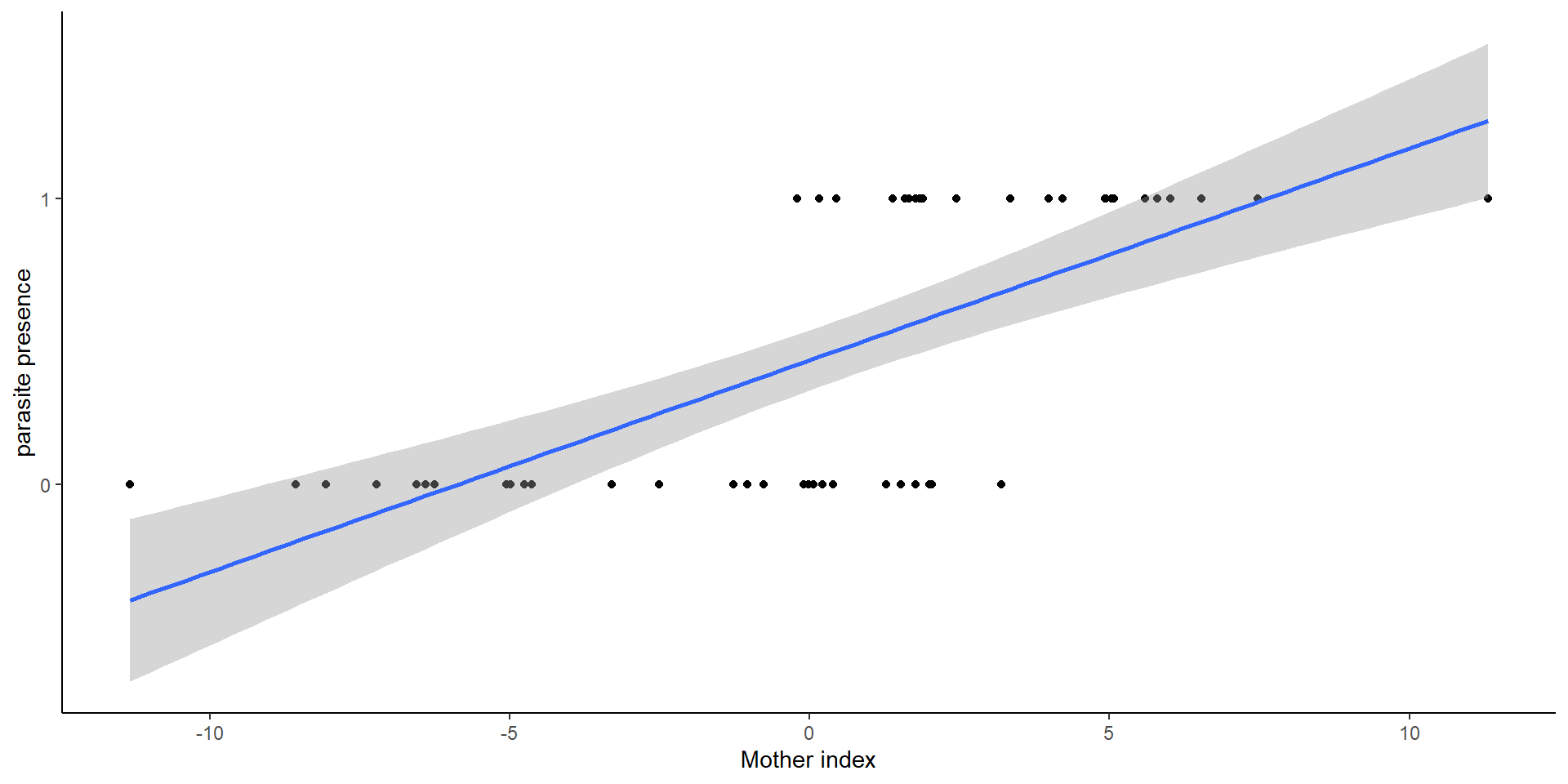
Generalized linear mixed effects models
Linear portion of the Mixed effects models have a deterministic and stochastic component
Count data
Binary data (1, 0)
GLMM’s
How can we run mixed effects models?
Easy way: Add random effects to the linear predictor, leading to generalized linear mixed effect models
Essentially, you have two sources of variation
One is normally dsitributed, the other one is distributed according to a different distribution
Example
A poisson glm:
\(log(\lambda) = \beta_0 + \beta_1x_i\)
\(y_i \sim Poisson(\lambda)\)
A glmm: Poisson-normal
\(log(\lambda) = (\beta_0+\gamma) + (\beta_1+\psi)x_{ij}\)
\(\gamma \sim N(0,\sigma_\gamma)\) , \(\psi\sim N(0,\sigma_\psi)\)
\(y_i \sim Poisson(\lambda)\)
Parameter interpretation
Not easy to interpret!
In random effects if we set all random effects to 0, then we estimate the mean for a “typical” individual
“typical means subject, site, individual, etc.”
The variances are in different distributions
Typical individual does not equal “population average response”
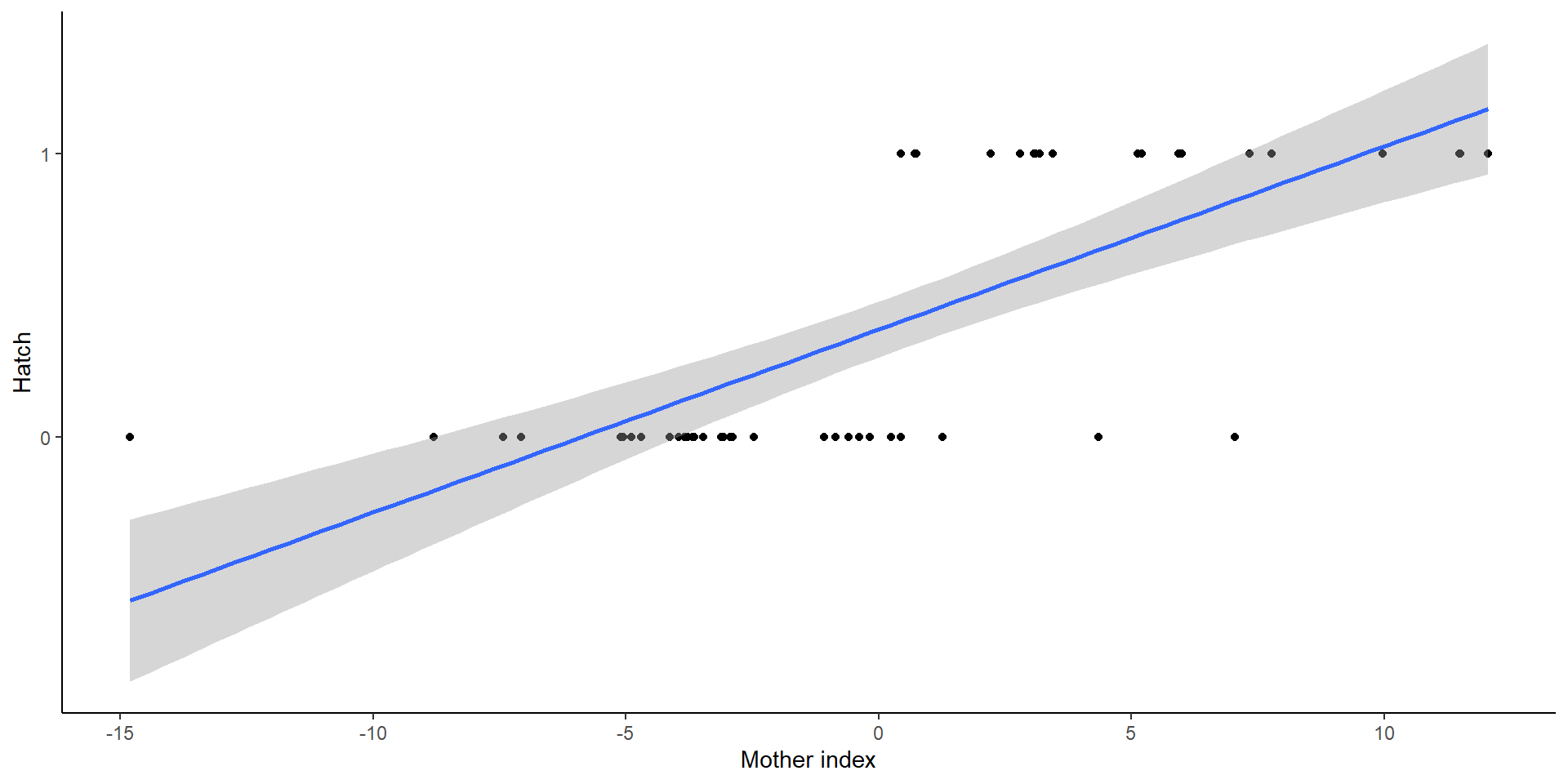
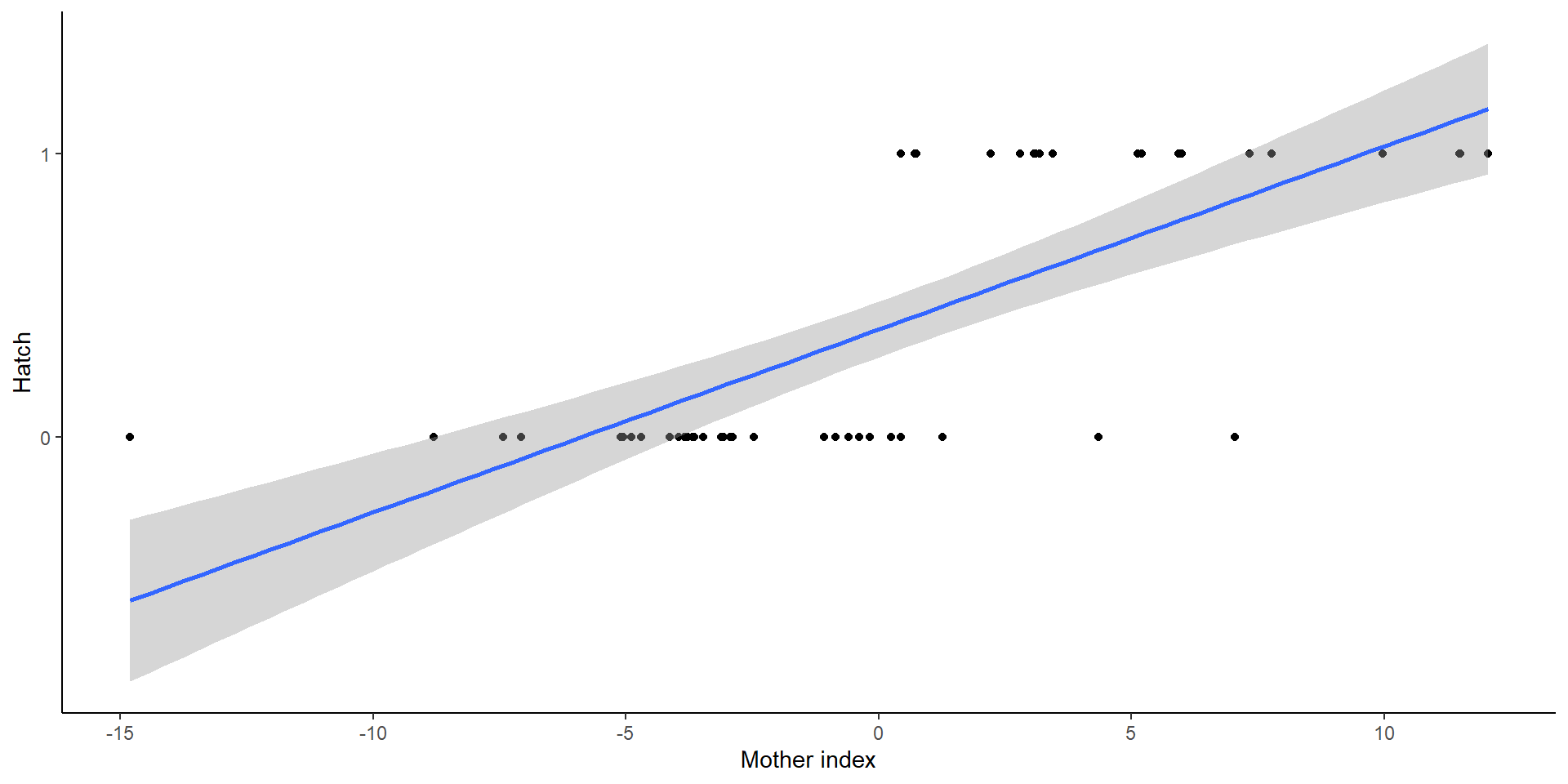
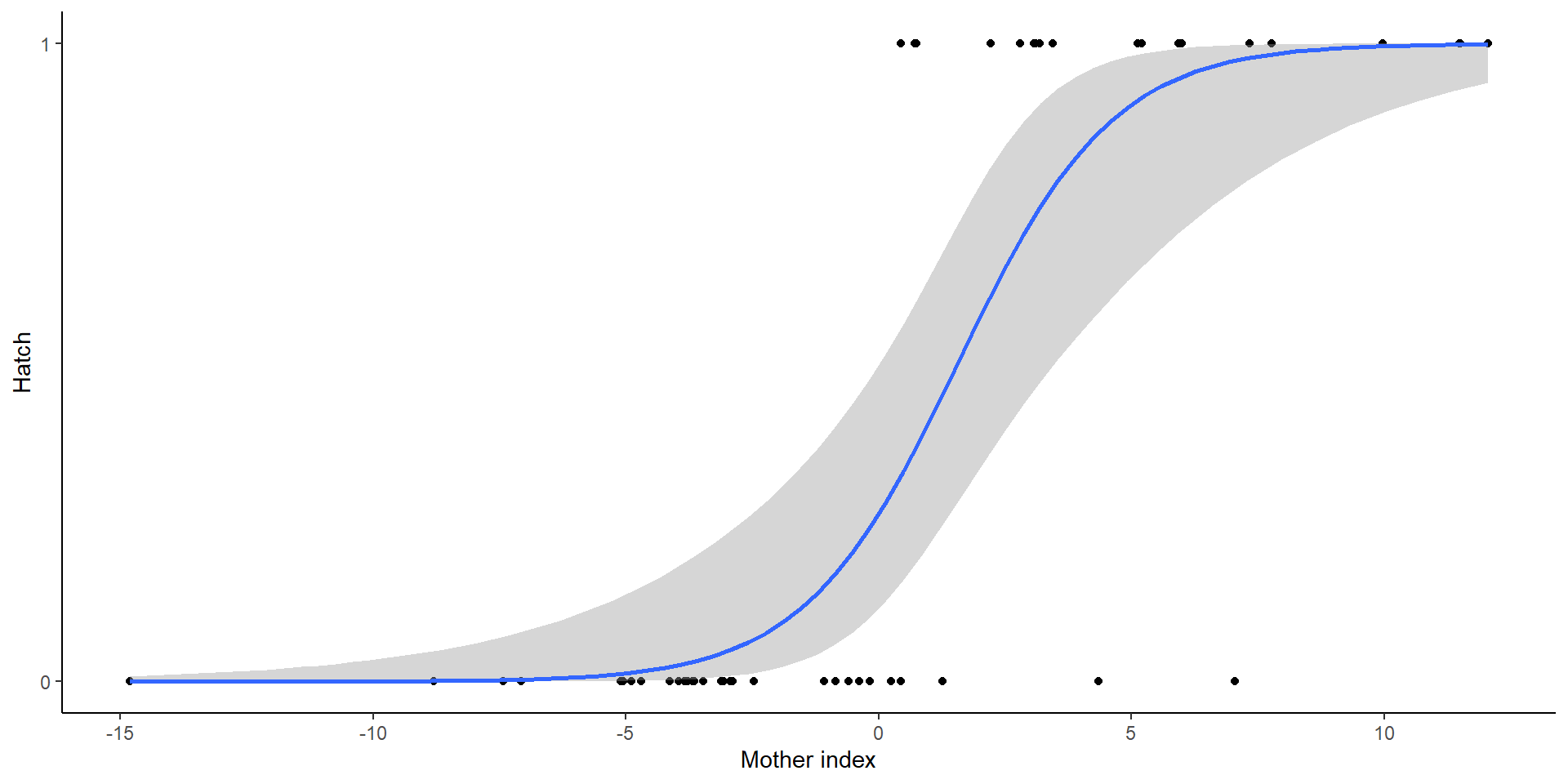
Example
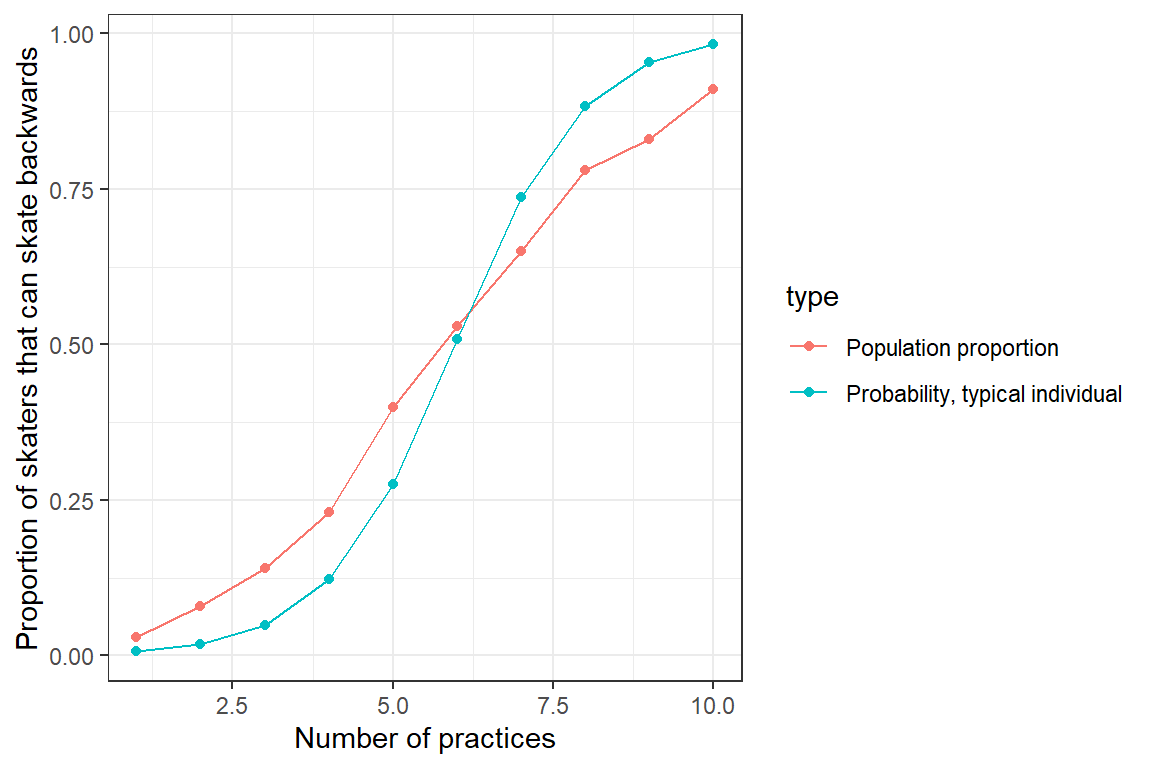
Both curves are not lining up
- Due to non-logit transformations (random effects are normal)
Interpreting data
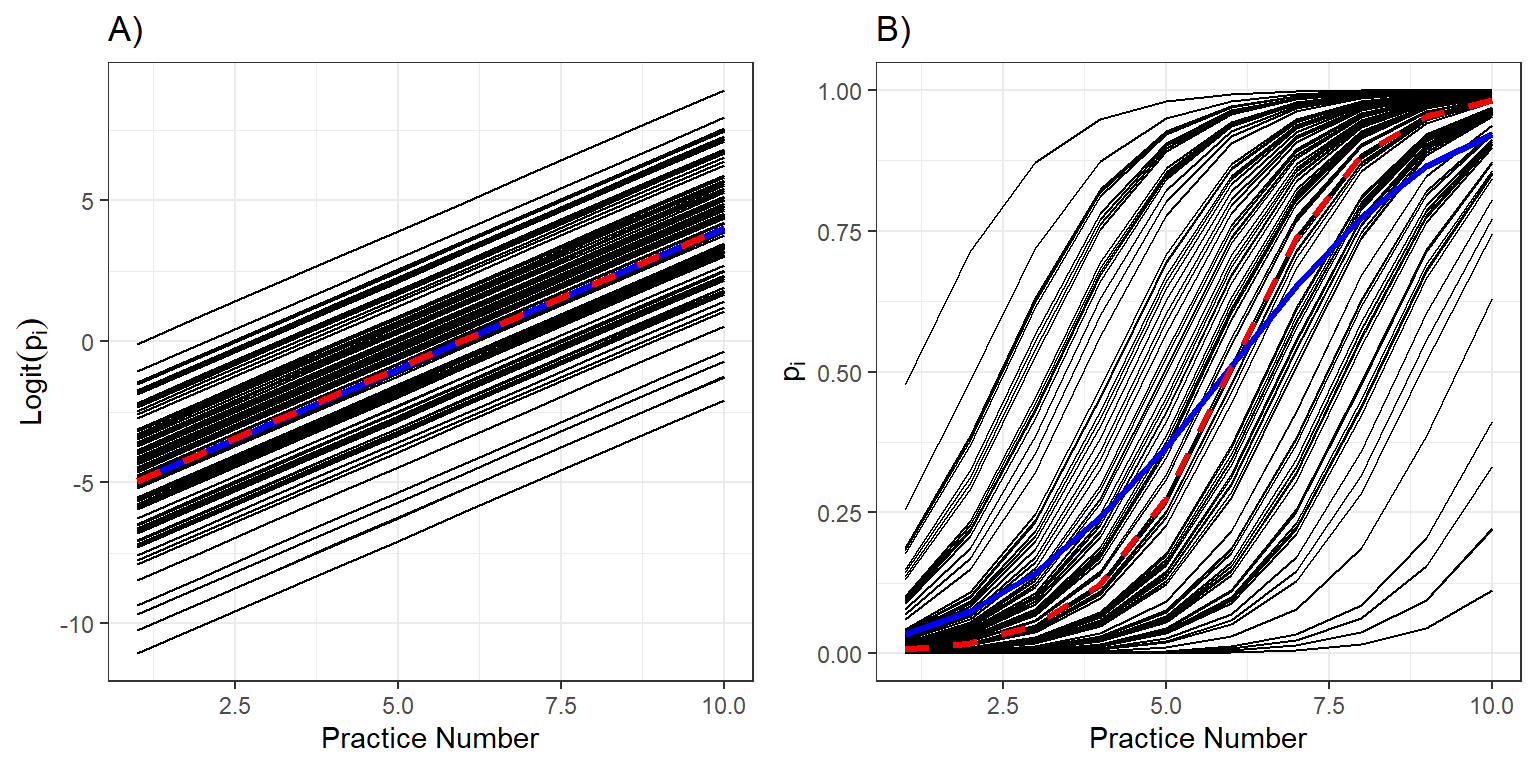
- Individual response curves (black), the response curve for a typical individual with random effects at zeroes, and the population mean response curve (blue) and on the logit and probability scales
In other words
A glmm: Poisson-normal
\(log(\lambda) = (\beta_0+\gamma) + (\beta_1+\psi)x_{ij}\)
\(\gamma \sim N(0,\sigma_\gamma)\) , \(\psi\sim N(0,\sigma_\psi)\)
\(y_i \sim Poisson(\lambda)\)
Because of non linearity if we set \(\gamma\) and \(\psi\) to 0’s, the end result will be different than if we get the mean of the overall response
Take away
If you do glmm’s be very careful about interpretation
- In general, transforming data can be risky
Solutions?
Package
GLMMadaptiveestimates marginal meansHow can we run mixed effects models?
Easy way: Add random effects to the linear predictor, leading to generalized linear mixed effect models
Hard way: Generalized Estimating Equations
Example
- Zuur, A., Ieno, E. N., Walker, N., Saveliev, A. A. & Smith, G. M. Mixed Effects Models and Extensions in Ecology with R. (Springer New York, 2009).
RIKZ data
RIKZ data
RIKZ institute
Inter-tidal area (AKA beach): 9
In each beach, 5 samples were taken
Response variable: macro-fauna richness (AKA number of species)
NAP: height of sampling station compared to mean tidal level
Exposure (index). Of multiple environmental conditions. Treated as categorical, as there are three levels
RIKZ data
flowchart TD A[RIKZ DATA] --> B(Beach 1) A --> C(Beach 2) A --> E(Beach 9) B --> F(site 1) B --> G(site 2) B --> H(site 3) B --> I(site 4) B --> J(site 5) C --> K(site 1) C --> L(site 2) C --> M(site 3) C --> N(site 4) C --> O(site 5) E --> P(site 1) E --> Q(site 2) E --> R(site 3) E --> S(site 4) E --> T(site 5)
Let’s work on this example
Objective
flowchart TD A[RIKZ DATA] --> B(Beach 1) A --> C(Beach 2) A --> E(Beach 9) B --> F(site 1) B --> G(site 2) B --> H(site 3) B --> I(site 4) B --> J(site 5) C --> K(site 1) C --> L(site 2) C --> M(site 3) C --> N(site 4) C --> O(site 5) E --> P(site 1) E --> Q(site 2) E --> R(site 3) E --> S(site 4) E --> T(site 5)
Exploring whether there is a relationship between richness and the two factors: NAP and exposure
We have an N of 45… but do we?
We have multiple potential alternatives: there is an effect of NAP, an effect of Exposure, an effect of both, of neither
Also… there are mixed effects
Random intercept, random slope, both? –> let’s wait to discuss this
Step 1: Read the data… there is a potential issue
- Step 2: decide our analysis! 🤔 Any ideas? any details?
The data
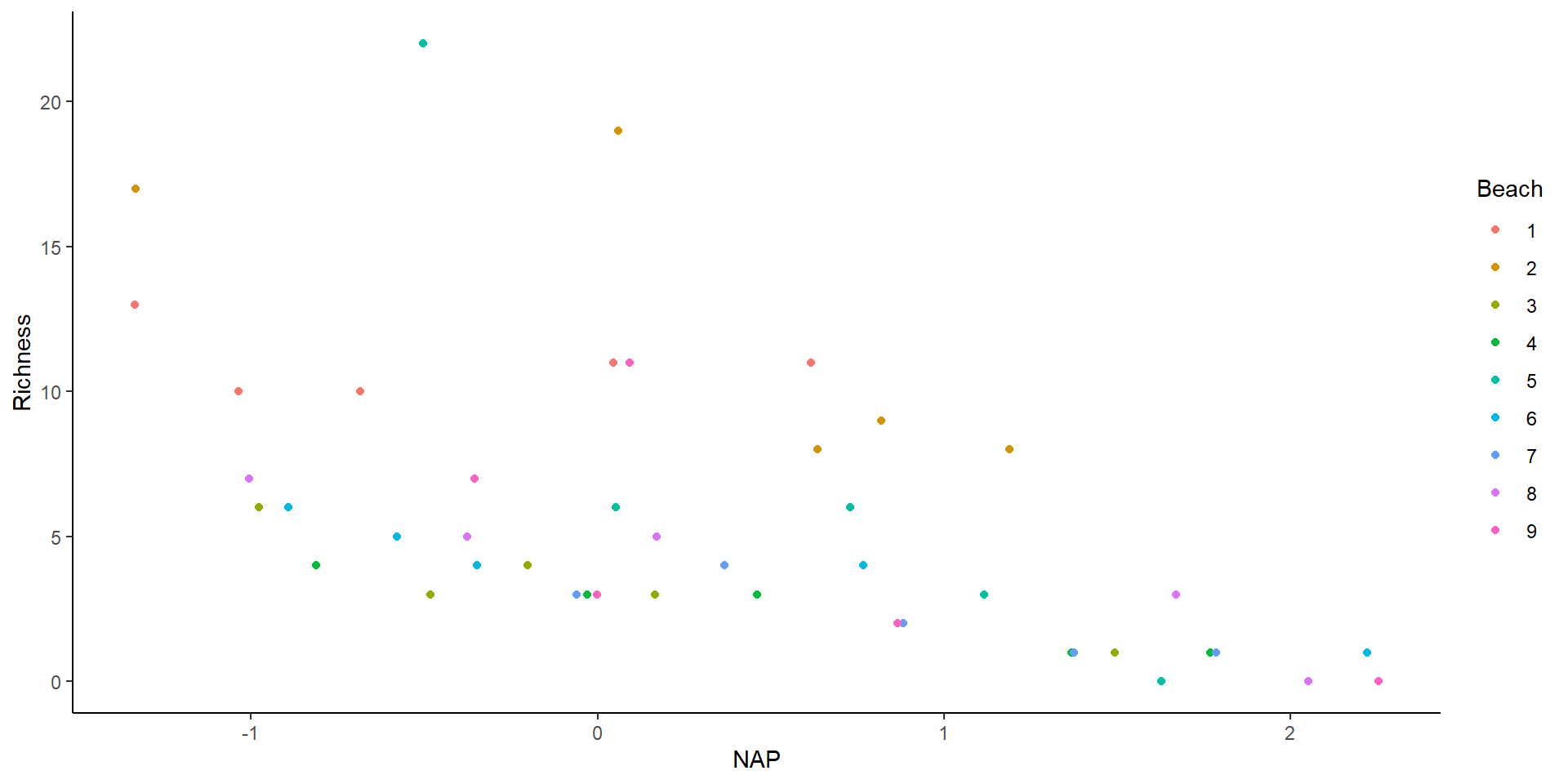
The data
There seems to be an effect of NAP…
how about… a random effect of beach?
step 3: how do we decide random effects?
step 3: random intercept? Or random slope? Or both? Or non
What do we do?
Exposure is categorical… so no random slope 😃
Model selection with random models
WE CANNOT TEST FIXED AND RANDOM EFFECTS AT THE SAME TIME
Test randomeffects first.
We use the “global” or saturated fixed model
How many models?
Use AIC
What model structure?
Model selection with random models
Four models. Run all with glmmTMB
Step 1: install the glmmTMB package
Model selection with random models
Fixed effects: NAP*Exposure
Family: Poisson
Model selection based on AICc:
K AICc Delta_AICc AICcWt Cum.Wt LL
Mod1 6 210.94 0.00 0.62 0.62 -98.36
Mod3 7 213.24 2.30 0.20 0.81 -98.10
Mod2 7 213.52 2.59 0.17 0.98 -98.25
Mod4 9 217.81 6.87 0.02 1.00 -97.33Are the AIC values the same?
Notes on AIC
I recommend using the AICcmodavg package and making a list!
Next step: let’s explore the model (factors!)
How do you explore the best model?
Let’s look at the summary
Analysis of Deviance Table (Type II Wald chisquare tests)
Response: Richness
Chisq Df Pr(>Chisq)
NAP 51.7328 1 6.359e-13 ***
Exposure 55.4525 2 9.092e-13 ***
NAP:Exposure 3.5604 2 0.1686
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Next step: let’s explore the model (factors!)
Analysis of Deviance Table (Type II Wald chisquare tests)
Response: Richness
Chisq Df Pr(>Chisq)
NAP 51.7328 1 6.359e-13 ***
Exposure 55.4525 2 9.092e-13 ***
NAP:Exposure 3.5604 2 0.1686
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Talk about Zuur’s approach here
Next step?
model1<-glm(Richness~NAP*Exposure, data = rikz, family = poisson)
model2<-glm(Richness~NAP+Exposure, data = rikz, family = poisson)
model3<-glm(Richness~NAP, data = rikz, family = poisson)
model4<-glm(Richness~Exposure, data = rikz, family = poisson)
model5<-glm(Richness~1, data = rikz, family = poisson)
modeltab<-AICcmodavg::aictab(list(model1,model2,model3,model4,model5))
modeltab
Model selection based on AICc:
K AICc Delta_AICc AICcWt Cum.Wt LL
Mod2 4 209.37 0.00 0.69 0.69 -100.19
Mod1 6 210.94 1.57 0.31 1.00 -98.36
Mod3 2 259.47 50.10 0.00 1.00 -127.59
Mod4 3 265.87 56.50 0.00 1.00 -129.64
Mod5 1 323.84 114.47 0.00 1.00 -160.88Evidence ratios

Burnham and Anderson
Evidence ratios
Evidence ratio between models 'Mod2' and 'Mod1':
2.19 - Raffle tickets
Last step: Model validation
Of the best model.
Other steps?
Check assumptions (if normal)